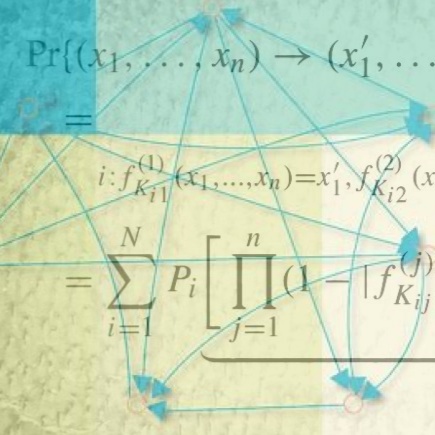
Probabilistic Boolean Networks are a class of models of genetic regulatory networks, first introduced in [1,2]. Our work has focused on the relationships between network structure and dynamics [6], steady-state analysis [5,8], and relationships to other model classes, such as Dynamic Bayesian Networks [7]. We have also worked on model inference from experimental data [9] and intervention strategies for driving the network towards desirable states [3,4]. This and the work of multiple other authors was included in the book on Probabilistic Boolean Networks [10] published by SIAM Press in 2010 with long-term colleague Dr. Edward Dougherty (Texas A&M). Recently, PBNs have been used, together with single cell expression data, for identifying the best intervention targets to induce transdifferentiation between two cell types [11].
Publications
- I. Shmulevich, E. R. Dougherty, S. Kim, W. Zhang, “Probabilistic Boolean Networks: A Rule-based Uncertainty Model for Gene Regulatory Networks,” Bioinformatics, Vol. 18, No. 2, pp. 261-274, 2002.
- I. Shmulevich, E.R. Dougherty, and W. Zhang, “From Boolean to probabilistic Boolean networks as models of genetic regulatory networks,” Proceedings of the IEEE, Vol. 90, No. 11, pp. 1778-1792, 2002.
- I. Shmulevich, E. R. Dougherty, W. Zhang, “Gene Perturbation and Intervention in Probabilistic Boolean Networks,” Bioinformatics, Vol. 18, No. 10, pp. 1319-1331, 2002.
- I. Shmulevich, E.R. Dougherty, and W. Zhang, “Control of stationary behavior in Probabilistic Boolean Networks by means of structural intervention,” Journal of Biological Systems, Vol. 10, No. 4, pp. 431-445, 2002.
- I. Shmulevich, I. Gluhovsky, R. Hashimoto, E. R. Dougherty, and W. Zhang, “Steady-State Analysis of Genetic Regulatory Networks Modeled by Probabilistic Boolean Networks,” Comparative and Functional Genomics, Vol. 4, No. 6, pp. 601-608, 2003.
- E. R. Dougherty and I. Shmulevich, “Mappings Between Probabilistic Boolean Networks,” Signal Processing, Vol. 83, No. 4, pp. 799-809, 2003.
- H. Lähdesmäki, S. Hautaniemi, I. Shmulevich, O. Yli-Harja, “Relationships Between Probabilistic Boolean Networks and Dynamic Bayesian Networks as Models of Gene Regulatory Networks,” Signal Processing, Vol. 86, No. 4, pp. 814-834, 2006.
- M. Brun, E. R. Dougherty, I. Shmulevich, “Steady-State Probabilities for Attractors in Probabilistic Boolean Networks,” Signal Processing, Vol. 85, No. 4, pp. 1993-2013, 2005.
- R. F. Hashimoto, S. Kim, I. Shmulevich, W. Zhang, M. L. Bittner, E. R. Dougherty, “Growing genetic regulatory networks from seed genes,” Bioinformatics, Vol. 20, No. 8, pp. 1241-1247, 2004.
- I. Shmulevich and E. R. Dougherty, Probabilistic Boolean Networks: The Modeling and Control of Gene Regulatory Networks, SIAM Press, 2009.
- B. Tercan, B. Aguilar, S. Huang, E. R. Dougherty, I. Shmulevich, “Probabilistic Boolean Networks Predict Transcription Factor Targets to Induce Transdifferentiation,” iScience, Vol. 25, No. 9, 104951, 2022.


 thorsson-shmulevich.isbscience.org/research/pbns/
thorsson-shmulevich.isbscience.org/research/pbns/